Following on the immunoprecipitation article for the study of protein-protein interactions, we will discuss the most commonly used technique for protein-DNA interactions, chromatin immunoprecipitation (ChIP), in this week’s Antibody Application series.
Our guest writer this week, Justyna Kulpa, recently completed her PhD in Molecular Biology at University of Montreal, and has extensive experience working with ChIP.
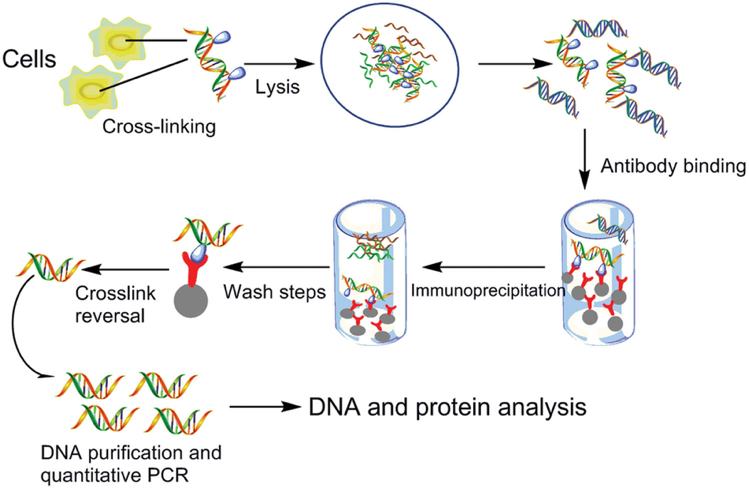
Image credit: Song, C., Zhang, S., and Huang, H. (2015). Choosing a suitable method for the identification of replication origins in microbial genomes. Front. Microbiol.
What Is Chromatin Immunoprecipitation?
Not only do proteins interact with one another, they can also interact with DNA. Chromatin immunoprecipitation (ChIP) is a technique that determines whether a protein of interest interacts with a specific DNA sequence. This technique is often used to study the repertoire of sites on DNA that are bound by particular transcription factors or by histone proteins, and to look at the precise genomic locations of various histone modifications (including acetylation, phosphorylation, or methylation).
How Does ChIP Work?
ChIP can be used to examine the presence of protein-DNA interaction at steady state, or to quantify changes in interaction at specific phases of the cell cycle, or following a treatment of interest. Protein and associated chromatin are temporarily cross-linked in live cells or tissues (using formaldehyde or UV) and sheared using enzymatic digestion or sonication to yield ~300-1000 bp fragments of DNA. The protein of interest, along with any associated DNA fragments, is immunoprecipitated from the cell debris using a specific antibody. The cross-link is then reversed and DNA fragments are purified. The amount of eluted DNA can be assessed through quantitative real-time PCR (qRT-PCR) using primers flanking the genomic locus of interest. DNA amplification is an indication of enrichment in binding of the protein of interest.
Modified ChIP Techniques
DNA fragments purified by ChIP can be utilized for a number of downstream analysis techniques. Furthermore, the basic ChIP protocol described above can be modified to answer additional biological questions.
- ChIP-on-chip: Genome-wide analysis of protein binding sites using microarray analysis of purified DNA fragments.
- ChIP-Seq: Genome-wide analysis of protein binding sites using deep sequencing of purified DNA fragments.
- Native ChIP: Omits the cross-linking step and uses micrococcal nuclease digestion to cut DNA at histone linkers to examine the DNA target of histone modifying proteins.
- ChIP-exo: Addition of an exonuclease digestion step to obtain increased resolution of protein binding sites, up to a single base pair.
- ChIA-PET (chromatin interaction analysis by paired-end tag sequencing): A technique that combines the principles of ChIP with chromosome conformation capture (3C) to detect long-range chromatin interactions mediated via a protein of interest.
- iChIP (indexing-first chromatin immunoprecipitation): A high-sensitivity technique that reduces the number of cells required for a ChIP experiment by initially barcoding total cellular chromatin.
- enChIP (engineered DNA-binding molecule-mediated chromatin immunoprecipitation): A technique which employs the CRISPR/Cas9 system to target specific genomic regions. A guide RNA complementary to the desired genomic region is expressed in combination with a tagged, enzymatically inactive Cas9 protein. ChIP is then performed using an antibody against the modified Cas9. This technique can help evaluate cis- and trans-interacting chromosomal looping events.
- RIP-Chip/RIP-Seq: Similar techniques used to analyze protein-RNA interactions.
Limitations of ChIP
As with all molecular biology techniques, ChIP is not without its own set of limitations. ChIP assays often yield low signals as compared with controls, leading to inconclusive data. The assay is limited to a resolution relative to the size of the DNA fragments generated following shearing, which makes it difficult to determine the exact binding site of a protein. While ChIP will infer the presence of a protein at a given genomic locus, it cannot determine functional significance of the protein’s binding at that DNA region. Cross-linking may additionally include proteins that transiently interact with DNA or DNA-binding proteins and have no functional significance. Similarly, DNA interactions of proteins with short residence time (as little as several seconds for some transcription factors) may not be fully captured. Additionally, interacting proteins may mask the epitope of the protein of interest. Finally, the ChIP technique is extremely dependent on the quality and specificity of the antibody employed and may not discriminate between different DNA-binding protein isoforms.
Important Factors to Consider
While the principle of the ChIP technique is quite simple, the actual execution of a ChIP experiment can be rather complex. Here are some important factors to consider when planning or trouble-shooting ChIP assays:
1. Garbage in, Garbage Out
The quality of the chromatin generated in ChIP can make the difference between an uninterpretable result and a quality data point. In particular, sonication is typically used to break chromatin down into shorter fragments, but sonicating for too long or too harshly can lead to denatured protein, or protein that is dissociated from DNA or whose antibody epitope has been destroyed. This step requires considerable optimization. Four key variables to consider are cell density, power, time and number of cycles. Be sure to keep chromatin on ice at all times, and limit pulses to under 30 seconds at a time to prevent protein denaturation from overheating.
2. Antibody Validation
While antibodies for western blotting can recognize denatured protein, ChIP requires recognition of the tertiary structure of a protein of interest and of an exposed epitope. ChIP-validated or ChIP-grade antibodies are recommended, but antibodies validated for immunoprecipitation (IP) or immunohistochemistry (IHC) experiments may work as well. Another consideration is the choice between monoclonal or polyclonal antibodies. While monoclonal antibodies have higher batch-to-batch consistency, they recognize a single epitope, which may be masked through binding to other proteins or buried within the tertiary structure of the protein. Polyclonal antibodies on the other hand represent a population of a number of different antibodies that recognize different epitopes. This reduces the probability that all specific epitopes are masked due to cross-linking and increases the chance of a positive result. Alternatively, if no good antibody candidates exist, it is possible to tag the protein of interest with a Myc, His or GST tag, for example, and use antibodies targeting the tag of choice.
3. Cell Number Considerations
It is also important to optimize the number of cells for your particular ChIP experiment. This will depend on the abundance of the target being studied. Histone modifications are quite abundant, and ChIPping for these marks generally requires less starting material than probing for transcription factors, which are significantly scarcer.
4. Cross-Linking Issues
Cross-linking is a time-sensitive step and should only be performed for short durations (~10 minutes). Prolonged cross-linking can reduce antigen availability and efficiency of the chromatin-shearing step. Excessive cross-linking may mask the epitope that is recognizable by your antibody, leading to a reduction in protein that can be pulled down.
5. Appropriate Controls
As for any experiment, controls are indispensable in ChIP to calculate enrichment in protein binding and to interpret the significance of a result. The following controls are recommended:
- IgG control (isotype-matched control immunoglobulin): The appropriate non-specific IgG is added instead of protein-specific antibody but at the same concentration, to give an indication of the assay background.
- Input control: Save 5-10% of your cell lysate before addition of antibodies to normalize for variations in starting material between consecutive experiments.
- Positive control locus: If possible, a DNA region where your protein of interest is known to bind should be examined. This will tell you if your ChIP experiment worked.
- Negative control locus: If possible, a DNA region where your protein of interest is not expected to bind should also be tested. This will tell you that your ChIP experiment is specific.
Let me know in the comment section below if you have specific questions regarding your ChIP experiment. Happy researching!
Editor’s Note: If you’re planning a ChIP experiment, be sure to take advantage of BenchSci’s technique filter to review and compare published ChIP data for your antibody search.
{{cta(‘fb70bc62-b226-4926-8c2b-732f72384343’)}}